ADVERTISEMENTS:
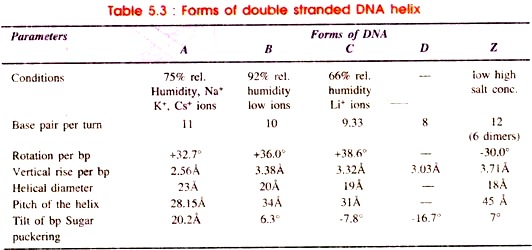
The most common form of DNA which has right handed helix and proposed by Watson and Crick is called B-form of DNA or B-DNA. In addition, the DNA may be able to exist in other forms of double helical structure. These are A and C forms of double helix which vary from B- form in spacing between nucleotides and number of nucleotides per turn, rotation per base pair, vertical rise per base pair and helical diameter (Table 5.3).
1. The B-Form of DNA (B-DNA):
Structure of B-form of DNA has been proposed by Watson and Crick. It is present in every cell at a very high relative humidity (92%) and low concentration of ions. It has antiparallel double helix, rotating clockwise (right hand) and made up of sugar- phosphate back bone combined with base pairs or purine-pyrimidine.
ADVERTISEMENTS:
The base pairs are perpendicular to longitudinal axis of the helix. The base pairs tilt to helix by 6.3°. The B-form of DNA is metabolically stable and undergo changes to A, C or D forms depending on sequence of nucleotides and concentration of excess salts.
2. The A-Form of DNA (A-DNA):
The A-form of DNA is found at 75% relative humidity in the presence of Na+, K+ or Cs+ ions. It contains eleven base pairs as compared to ten base pairs of B-DNA which tilt from the axis of helix by 20.2°. Due to this displacement the depth of major groove increases and that of minor groove decreases. The A-form is metastable and quickly turns to the D-form.
3. The C-Form DNA (C-DNA):
The C-form of DNA is found at 66% relative humidity in the presence of lithium (Lit+) ions. As compared to A-and B-DNA, in C-DNA the number of base pairs per turn is less i.e. 28/3 or 9 1/3. The base pairs show pronounced negative tilt by 7.8°.
4. The D-Form of DNA (D-DNA):
The D-form of DNA is found rarely as extreme vanants. Total number of base pairs per turn of helix is eight. Therefore, it shows eight-fold symmetry. This form is also called poly (dA-dT) and poly (dG-dC) form. There is pronounced negative tilt of base pairs by 16.7° as compared to C form i.e. the base pairs are displaced backwardly with respect to the axis of DNA helix.
5. The Z-Form of DNA (Z-DNA) or Left Handed DNA:
ADVERTISEMENTS:
In 1979, Rich and coworkers at MIT (U.S.A.) obtained Z-DNA by artificially synthesizing d (C-G) 3 molecules in the form of crystals. They proposed a left handed (synistral) double helix model with zig-zag sugar-phosphate back bone running in antiparallel direction.
Therefore, this DNA has been termed as Z-DNA. The Z-DNA has been found in a large number of living organisms including mammals, protozoans and several plant species.
There are several similarities with B-DNA in having:
(i) Double helix,
(ii) Two antiparallel strands, and
(iii) Three hydrogen bonds between G-C pairing.
In addition, the Z-DNA differs from the B-DNA in the following ways:
(a) The Z-DNA has left handed helix, while the B-DNA has right handed helix.
(b) The Z-DNA contains zig-zag sugar phosphate back bone as compared to regular back bone of the B-DNA.
ADVERTISEMENTS:
(c) The repeating unit in Z-DNA is a dinucleotide due to alternating orientation of sugar residues, whereas in B-DNA the repeating unit is a mononucleotide, and sugar molecules do not have the alternating orientation.
(d) In the Z-DNA one complete turn contains 12 base pairs of six repeating dinucleotide, while in B-DNA one full turn consists of 10 base pairs i.e. the 10 repeating units.
(e) Due to the presence of high number (12) of base pairs in one turn of Z-DNA, the angle of twist per repeating unit i.e. dinucleotide is 60° as compared to 36° of B-DNA molecule.
(f) In Z-DNA the distance of twist making one turn of 360° is 45Å as against 34Å in B-DNA.
ADVERTISEMENTS:
(g) The Z-DNA has fewer diameters (18Å) as compared to the B-DNA (20Å diameter).
6. Single Stranded (ss) DNA:
Almost all the organisms contain double stranded DNA except a few viruses such as bacteriophage φ × 174 which consists of single stranded circular DNA. It becomes double stranded only at the time of replication.
The differences of ssDNA from the dsDNA are as below:
(a) The dsDNA absorbs wavelength 2600 Å of ultra violet light constantly from 0 to 80°C, thereafter rise sharply, whereas in ssDNA absorption of UV light increases steadily from 20° to 90°C.
ADVERTISEMENTS:
(b) The dsDNA resists the action of formaline due to closed reactive site, while the ss DNA does not resist it due to exposed reactive sites.
(c) Base pair composition in dsDNA is equal i.e. A=T and G=C, in ssDNA the composition of A, T, G, C is in proportion of 1:1.33:0.98:0.75.
(d) The dsDNA always remains in linear helical form, while the ssDNA remains in circular form; however, it becomes double stranded only during replication (i.e. replicative form).
7. Circular and Super Helical DNA:
Almost in all the prokaryotes and a few viruses, the DNA is organised in the form of closed circle. The two ends of the double helix get covalently sealed to form a closed circle. Thus, a closed circle contains two unbroken complementary strands. Sometimes one or more nicks or breaks may be present on one or both strands, for example DNA of phage PM2 (Fig. 5.7 A).
Besides some exceptions, the covalendy closed circles are twisted into super helix or super coils (Fig.5.7 B) and is associated with basic proteins but not with histones found complexed with all eukaryotic DNA.
This histone like proteins appear to help the organization of bacterial DNA into a coiled chromatin structure with the result of nucleosome like structure, folding and super coiling of DNA, and association of DNA polymerase with nucleoids. Several histones like DNA binding proteins have been described in bacteria (Table 5.4).
These nucleoid-associated proteins include HU proteins, IHF, protein H1, Fir A, H-NS and Fis. In archaeobacteria (e.g. Archaea) the chromosomal DNA exists in protein-associated form. Histone like proteins has been isolated from nucleoprotein complexes in Thermoplasma acidophilurn and Halobacterium salinanim.
Thus, the protein associated DNA and nucleosome like structures are detected in a variety of bacteria. If the helix coils clockwise from the axis the coiling is termed as positive or right handed coiling. In contrast, if the path of coiling is anticlockwise, the coil is called left handed or negative coil.
Table 5.4 : Histone-like proteins of E. coli
The two ends of a linear DNA helix can be joined to form each strand continuously. However, if one of ends rotates at 360° with respect to the other to produce some unwinding of the double helix, the ends are joined resulting in formation of a twisted circle in opposite sense i.e. opposite to unwinding direction.
ADVERTISEMENTS:
Such twisted circle appears as 8 i.e. it has one node or crossing over point. If it is twisted at 720° before joining, the resulting super helix will contain two nodes (Fig. 5.7B).
The enzyme topoisomerases alter the topological form i.e. super coiling of a circular DNA molecule. Type I topoisomerases (e.g. E.coli Top A) relax the negatively super coiled DNA by breaking one of the phosphodiester bonds in dsDNA allowing the 3′-OH end to swivel around the 5′-phosphoryl end, and then resealing the nicked phosphodiester backbone.
Type II topoisomerases need energy to unwind the DNA molecules resulting in the introduction of super coils. One of type II isomerases, the DNA gyrase, is apparently responsible for the negatively super coiled state of the bacterial chromosome. Super coiling is essential for efficient replication and transcription of prokaryotic DNA.
The bacterial chromosome is believed to contain about 50 negatively super coiled loops or domains. Each domain represents a separate topological unit, the boundaries of which may be defined by the sites on DNA that limit its rotation.